In a (somewhat) recent blog post entitled “Is there a crisis in Anolis taxonomy?”, Julian Velasco invited discussion on a perceived decline in the number of new anole taxonomists. While it was a fun look at the dynamics of anole taxonomy over time, I couldn’t help but feel like there is a more pressing taxonomic crisis going on right now, and it affects many of the researchers that frequent this blog.
I fear too many species of Anolis are being described based on questionable evidence. While this problem is not unique to anoles (a common term for it is “taxonomic inflation”; Isaac et al. 2004), a number of recently described anole species may be the result of overzealous taxonomic splitting. I will give some examples below and then briefly discuss two lines of evidence that I believe are often used to divide species inappropriately. Before I do so, it’s worth stating up front that I’ll focus on the work of Dr. Gunther Köhler and colleagues. This shouldn’t be surprising, as Dr. Köhler is the most prolific living describer of anole species. The following criticisms should not be seen as personal, as Köhler is not unique on any of the points I discuss below. But with many cryptic species described or resurrected over the past 10-15 years, his work has the largest impact on anole taxonomy and the science that depends on it.
I’ll start with the revision of the Anolis tropidonotus complex published in Mesoamerican Herpetology (Köhler et al. 2016). Below I provide a quick breakdown of the paper. I hope that others will contribute their own views on this work in the comments. The A. tropidonotus group is one that I am well-acquainted with, having spent months of field time collecting individuals across the distribution of the group. Köhler et al. (2016) raise a subspecies (A. tropidonotus spilorhipis) to species status while describing two new species, A. wilsoni and A. mccraniei. Unfortunately, the data presented–morphology and DNA–do not appear to strongly support the recognition of any new species level taxa. I argue that the inference of four species within A. tropidonotus sensu lato should require stronger evidence than that presented.
The authors sequenced 16S mitochondrial DNA for molecular analyses and present a consensus tree from Bayesian analyses of these data. This tree recovers four well-supported and geographically circumscribed mtDNA haplotype clades that correspond with the four new species. A table following the tree reveals the genetic distances between putatively new species topped out at 4.5%. This level of mitochondrial divergence is significantly less than intraspecific variation observed in other anoles (Malhotra & Thorpe 2000; Thorpe & Stenson 2003; Ng & Glor 2011). Moreover, Köhler et al.’s (2016) sampling map reflects sparse sampling of molecular data.
Based on Figure 3, morphology (other than perhaps hemipenes, which I discuss below) does not provide any support for delimitation of those populations characterized by distinct mtDNA haplotypes. The dewlap differences reported are slight and appear to fall within the type of variation observed within and among other populations of species in this group (see photos at the top of this post for an example of two spilorhipis males that came from the same locality; photos courtesy Luke Mahler). Bottom line–we see several populations with mitochondrial haplotypes that cluster together geographically with little to no morphological evidence for divergence.
The phylogenetic and morphological patterns displayed in Köhler et al. (2016) are consistent with patchy sampling of a widespread and continuously distributed species with potentially locally-adapted populations. The authors cite “the high degree of genetic distinctiveness… as evidence for a lack of gene flow, and conclude that these four lineages represent species-level units” (Köhler et al. 2016). This assumption is questionable, as researchers have long known of the pitfalls of using mtDNA to determine gene flow (Avise et al. 1983; Avise et al. 1984; Funk & Omland 2003) and supporting evidence from morphology is lacking. The different hemipenial types represent the strongest evidence for recognizing the lineages mtDNA haplotype groups. Below I will discuss the utility of those traits for species delimitation.
Finally, the authors did not compare their purported new tropidonotus-like species to Anolis wampuensis, a morphologically indistinguishable (McCranie & Kohler 2015) form that is potentially codistributed with the new species A. mccraniei. This should have been done to avoid the possibility that A. wampuensis is conspecific with one of the newly named forms.
Another example of taxonomic inflation in Anolis is from a 2014 monograph in Zootaxa (Köhler et al. 2014). I will focus on the treatment of the Anolis nebuloides group, which exemplifies the problems of this monograph and exhibits some of the issues I’ve seen repeated in species descriptions of late. In this paper, the authors use two mtDNA data sets: 16S and CO1. They present both trees for the nebuloides group; each tree is weakly supported and highly uncertain (below). The species are ultimately divided up on the basis of the molecular evidence.
As you can see on the map, there are five clusters of localities that appear to be separated by significant distances. These clusters correspond to each of the recognized species, and to OTUs in the phylogenetic trees. A reader unfamiliar with the region might interpret each species as a montane isolate, a common scenario involving species groups adapted to high elevations. However, this interpretation would be incorrect. Each of those clusters corresponds to a major road system that was sampled by the authors. In reality, the distribution of this group is likely to be continuous (or very nearly so) from the type locality of Anolis megapholidotus in Guerrero to at least the area in central Oaxaca where “zapotecorum” is reported. I have sampled within several of the putative gaps and have found populations of nebuloides-like anoles present at every single location I have checked within the outer boundaries of the distribution of the group, including very low elevations that would allow population connections between the sampled clusters. The authors should be aware of the elevational range of the group: they include on their map the type locality of the synonymized species Anolis simmonsi (=Anolis nebuloides), which is near 100 m elevation.
The authors cite the molecular evidence for splitting the group. The extent of the molecular evidence is that spatially proximal haplotypes cluster together in the authors’ phylogenetic analyses, which is exactly what one would expect from sparse sampling of a continuously distributed species. In other words, isolation by distance would explain their results perfectly, and sampling localities in-between could erase the artifactual pattern that the clusters are distinct evolutionary lineages. Once again, morphology does not corroborate the species delimited by the authors’ interpretation of the molecular data.
The tropidonotus and nebuloides anoles are only a couple examples. A careful reviewer might have raised some issues over the descriptions mentioned above, as well as many others. I suspect that reviewers may not be anole experts and therefore are unaware of the nuances of anole systematics. An example of this would be the relevance of a “lock-and-key” mechanism for reproductive isolation in the group. Generally, “lock and key” assumes differences in genital morphology evolved as a form of reproductive isolation between species (Shapiro & Porter 1989; Arnqvist 1998). While it is well-documented in some groups (especially many insects), we have yet to see published work that suggests an important role for “lock-and-key” reproductive isolation with respect to hemipenial morphology in anoles, and the literature is littered with examples that do not follow the expected pattern of isolation (Shapiro & Porter 1989).
What appears to be happening is that some authors are essentially defining species as the smallest group they can diagnose according to hemipenes or mitochondrial divergence/clustering alone. This is a dangerous way to approach taxonomy, as many species have diagnosable populations that freely share genetic material (Thorpe & Stenson 2003; Ng & Glor 2011; Ng et al. 2016). There is also a precedent for mtDNA markers misinforming researchers on the extent of gene flow between anole populations (Thorpe et al. 2008; Ng & Glor 2011). When dealing with putative cryptic species, an accurate assessment of gene flow is critical. And without demonstrating some level of reproductive isolation, the populations should not be recognized as species.
On that note: care should be taken when a single locus is used as the main line of evidence for splitting a species, especially if that locus is mtDNA. It’s become clear that mtDNA (or any single locus) can be misleading when trying to determine whether populations are reproductively isolated (Funk & Omland 2003; Petit & Excoffier 2009). In the case of mtDNA, this may be related to the male-biased nature of anole dispersal and mitochondria being a matrilineally inherited trait (Thorpe et al. 2008). Regardless, there are several examples of mitochondrially divergent populations of anoles having significant gene flow between them (Thorpe et al. 2008; Ng & Glor 2011), which should preclude species designations based on morphological differences the populations might have. Mitochondrial divergence of over 10% has been observed in multiple population pairings, yet extensive ongoing nuclear gene flow has ruled out the possibility that these mitochondrial lineages represent valid species (Thorpe et al. 2008; Ng & Glor 2011). Had those authors relied primarily on mtDNA divergence to delimit species, flawed conclusions would have resulted despite morphological distinctiveness of some populations. If adding nuclear DNA evidence is not an option, mtDNA evidence should be strengthened by other traits that corroborate splits (such as dewlap color divergence, which has a well-documented history of usefulness in species delimitation in anoles; Losos 2009; Glor & Laport 2012) and should never stand alone.
Hemipenial variation, in particular, has been cited as the single diagnostic trait for many recently described anole species. However, to date, we have no evidence that divergence in hemipenial traits between populations leads to reproductive isolation, which would be necessary for hemipenial traits to be used in this way. In fact, the evidence is building against such a claim. As many of the readers of this blog are aware, hemipenes were recently found to evolve faster within anoles than any other trait (Klaczko et al. 2015). This result is consistent with other studies in animals that show rapid divergent evolution of male genitalia (Arnqvist 1998; Eberhard 2009). When dealing with such fast evolving traits, we should expect to see hemipenial polymorphism within species just as we see polymorphism in scale traits, etc. In fact, I and others have found exactly that–in a recently accepted manuscript, we report on hemipenial polymorphism within Anolis sericeus (Lara-Tufiño et al. 2016). Work from Dr. Köhler’s lab has shown one species diagnosed solely by hemipenial morphology to be invalid (Anolis osa; Köhler et al. 2010; Köhler et al. 2012). My own work on the A. sericeus group (in prep.) stands in strong contrast to the results of Köhler and Vesely (2010), which used hemipenes to divide up the group. Phillips et al.’s (2015) work on the A. humilis group showed that a species diagnosed solely by hemipenial structure (A. quaggulus) was found not to warrant species status.
Other similarly diagnosed species may await a similar fate. Anolis cryptolimifrons, for example, was diagnosed from A. limifrons only in hemipenial structure and is geographically surrounded by A. limifrons (Köhler & Sunyer 2008). Pending evidence of minor hemipenial differences causing reproductive isolation, it seems prudent to regard species such as A. cryptolimifrons with skepticism. A more parsimonious interpretation simply acknowledges the presence of hemipenial polymorphism in the species.
A final note on hemipenes and sampling: many of the species descriptions that utilize hemipenial morphology to justify taxonomic splits include relatively few examined samples (population-wise and overall number of individuals). For instance, large gaps exist where hemipene morphotypes are assumed in Köhler and Vesely (2010; see map below)–the authors did not include samples from a large portion of the group’s distribution (hemipenial sampling shown by filled in shapes in the map). This practice is less important in descriptions where hemipenes are unused or ancillary evidence, but critical in papers like Köhler and Vesely (2010) where hemipenial structure is the primary diagnostic trait used to identify purported species. In some papers–including the tropidonotus work discussed above (Köhler et al. 2016)–detailed information on the number of hemipenes sampled per locality is lacking, so we are unable to judge the distribution of this trait at all. These data are important when a fast-evolving trait such as hemipenes is cited as primary evidence for species status.
In the future, we may find cases where divergent hemipenes restrict gene flow between closely-related populations. But given current evidence, differences in hemipenial morphology should not be accepted as the primary evidence to delimit species.
I have described just a few cases of taxonomic inflation in anoles. There are many others that have occurred over the past 15 years. This influx of potentially illegitimate names poses real challenges for anole researchers. If your research depends on accurate diversity estimates in clades or accurate distributions for species units, results can be compromised. Conservation decisions become muddled, and even getting permits to do ecological work can become more problematic due to the fact that many of the new species mentioned in this post are believed to have small distributions and considered endangered by the authors (Köhler et al. 2014; McCranie and Köhler 2015; Köhler and Hedges 2016). When describing cryptic species in continuously distributed groups like nebuloides and tropidonotus, it is vital to demonstrate a lack of gene flow before changing the taxonomic status of populations. I hope that more rigorous testing for species boundaries will be provided in future taxonomic overhauls, as these changes have real consequences for other anole researchers. An easy fix to this problem would be to encourage the recognition of subspecies, as Dr. Hillis has suggested before on this blog. Recognizing taxonomic diversity below species level can be plenty useful for a plethora of research topics and is less harmful to other scientific ventures. Another option would be to treat new discovered mitochondrial haplotype clades as “Unconfirmed Candidate Species” (Padial et al. 2010) pending evidence of reproductive isolation.
Let’s hear your thoughts and comments!
Literature Cited
Arnqvist, G. (1998) Comparative evidence for the evolution of genitalia by sexual selection. Nature 393:784-786.
Avise, J.C., J.F. Shapira, S.W. Daniel, C.F. Aquadro, R.A. Lansman (1983) Mitochondrial differentiation during the speciation process in Peromyscus. Molecular Biology and Evolution 1983:38-56.
Avise, J.C., J.E. Neigel, J. Arnold (1984) Demographic influences on mitochondrial DNA lineage survivorship in animal populations. Journal of Molecular Evolution 20:99-105.
Eberhard, W.G. (2009) Evolution of genitalia: theories, evidence, and new directions. Genetica 138:5-18.
Funk, D.J., K.E. Omland (2003) Species-level paraphyly and polyphyly: frequency, causes, and consequences, with insights from animal mitochondrial DNA. Annual Review of Ecology, Evolution, and Systematics 34:397-423.
Glor, R.E., R.G. Laport (2012) Are subspecies of Anolis lizards that differ in dewlap color and pattern also genetically distinct? A mitochondrial analysis. Molecular Phylogenetics and Evolution 2012:255-260.
Isaac, N.J.B., J. Mallet, G.M. Mace (2004) Taxonomic inflation: its influence on macroecology and conservation. Trends in Ecology and Evolution 2004:464-469.
Köhler, G., S.B. Hedges (2016) A revision of the green anoles of Hispaniola with description of eight new species (Reptilia, Squamata, Dactyloidae). Novitates Caribaea 9:1-135.
Köhler, G., J. Sunyer (2008) Two new species of anoles formerly referred to as Anolis limifrons (Squamata, Polychrotidae). Herpetologica 2008:92-108.
Köhler, G., M. Vesely (2010) A revision of the Anolis sericeus complex with the resurrection of A. wellbornae and the description of a new species (Squamata, Polychrotidae). Herpetologica 66:207-228.
Köhler, G., D.M. Dehling, J. Köhler (2010) Cryptic species and hybridization in the Anolis polylepis complex, with the description of a new species from the Osa Peninsula, Costa Rica (Squamata, Polychrotidae). Zootaxa 2718:23-38.
Köhler, G., R. Gomez Trejo Perez, C.B.P. Petersen, F.R. Mendez de la Cruz (2014) A revision of the Mexican Anolis (Reptilia, Squamata, Dactyloidae) from the Pacific versant west of the Isthmus of Tehuantepec in the states of Oaxaca, Guerrero, and Puebla, with the description of six new species. Zootaxa 3,862:1-210.
Köhler, G., J.H. Townsend, C.B.P. Petersen (2016) A taxonomic revision of the Norops tropidonotus complex (Squamata, Dactyloidae), with the resurrection of N. spilorhipis (Alvarez del Toro and Smith, 1956) and the description of two new species. Mesoamerican Herpetology 3:8-41.
Köhler, J., M. Hahn, G. Kohler (2012) Divergent evolution of hemipenial morphology in two cryptic species of mainland anoles related to Anolis polylepis. Salamandra 48:1-11.
Klaczko, J., T. Ingram, J. Losos (2015) Genitals evolve faster than other traits in Anolis lizards. Journal of Zoology 295:44-48.
Lara-Tufino, J.D., A. Nieto Montes de Oca, A. Ramirez Bautista, L.N. Gray (In press) Resurrection of Anolis ustus Cope, 1864 from synonymy with Anolis sericeus Hallowell, 1856 (Squamata, Dactyloidae). Zookeys 619:147-162.
Losos, J. (2009) Lizards in an evolutionary tree: ecology and adaptive radiation of anoles. Vol. 10. University of California Press.
Malhotra, A., R.S. Thorpe (2000) The dynamics of natural selection and vicariance in the Dominican anole: patterns of within-island molecular and morphological divergence. Evolution 54:245-258.
McCranie, J.R., G. Köhler (2015) The anoles of Honduras. Bulletin of the Museum of Comparative Zoology, Special Publications Series 1:1-292.
Ng, J., R.E. Glor (2011) Genetic differentiation among populations of a Hispaniolan trunk anole that exhibit geographical variation in dewlap colour. Molecular Ecology 20:4302-4317.
Ng, J., A.G., Ossip-Klein, R.E. Glor (2016) Adaptive signal coloration maintained in the face of gene flow in a Hispaniolan Anolis lizard. BMC Evolutionary Biology 16:193.
Padial, J.M., A. Miralles, I. De La Riva, M. Vences (2010) The integrative future of taxonomy. Frontiers in Zoology 7:16.
Petit, R.J., L. Excoffier (2009) Gene flow and species delimitation. Trends in Ecology and Evolution 24:386-393.
Phillips, J.G., J. Deitloff, C. Guyer, S. Huetteman, K.E. Nicholson (2015) Biogeography and evolution of a widespread Central American lizard species complex: Norops humilis, (Squamata, Dactyloidae) 15:143.
Shapiro, A.M., A.H. Porter (1989) The lock-and-key hypothesis: evolutionary and biosystematic interpretation of insect genitalia. Annual Review of Entomology 34:231-245.
Thorpe, R.S., A.G. Stenson (2003) Phylogeny, paraphyly, and ecological adaptation of the colour and pattern in the Anolis roquet complex on Martinique. Molecular Ecology 12:117-132.
Thorpe, R.S., Y. Surget-Groba, H. Johansson (2008) The relative importance of ecology and geographic isolation for speciation in anoles. Philosophical Transactions of the Royal Society B 363:3071-3081.
- Dewlap Size and Seasonality in Mexican Anoles - May 18, 2020
- Mexican Anole Primer, Part 1: Smooth Ventral Scales - October 9, 2018
- Vanzolini’s Anole Video - April 20, 2017









Peter Uetz
As Levi indicated (but not mentioned explicitly), Gunther Köhler has provided some evidence that even species with different hemipenial morphologies appear to hybridize, e.g. A. osa and A. polylepis (Köhler et al. 2012, loc. cit.).
Other cases of doubtful anoles have been discussed in AA by Steve Poe (https://www.anoleannals.org/wp-content/uploads/2016/06/Poe.-Anoles-of-Honduras-review.-2016.pdf).
If you are aware of such instances, please let me know. I am happy to add a comment to these species in the Reptile Database (several will appear in the next release).
Ambika Kamath
Thanks for this really important post, Levi. I’ve had a similar (but far less comprehensive) post drafted for ages, and I’ve posted some excerpts from that below. I hope that my points will complement the ones you’ve made in this post.
I approach this whole debate not as a taxonomist, but as a behavioural ecologist who usually pays more attention to within-population variation than to whether or not two populations belong to the same species. But I think that within-population variation has important consequences for the way in which many new lizard species are described these days. My main gripe is simple: individuals within populations vary, and how much they vary affects how well we can tell populations apart. For species defined on the basis of traits that vary, it matters whether or not the ranges of these traits overlap, and how much they overlap by. If within-population variation equals or exceeds the difference between population means for these traits, then they aren’t useful in distinguishing between these populations, and one cannot describe these populations as different species based on these traits. This clearly seems to be the case for the dewlaps of Anolis tropidonotus as you describe, and I’ve noted something similar in Sitana as well (e.g. https://www.anoleannals.org/2015/02/14/two-new-species-of-fan-throated-lizards-from-sri-lanka/)
The absence of morphological differences between populations of course does not necessarily mean the populations in question are not different species, but it does mean that the evidence for their distinctness is weakened. Using the Generalized Lineage Concept for defining species based on multiple lines of evidence (morphological, reproductive, phylogenetic) is certainly biologically more meaningful than strict adherence to any one species concept, but it isn’t clear to me how strong each line of evidence needs to be before you define a species, and what the consequences are of the weakening of one line of evidence.
Whether or not we like it, species definitions influence how all of us non-taxonomists structure our research. The way in which species are defined can thus legitimize (or de-legitimize) certain avenues of research. Let’s imagine a species that’s split up based in large part on geographic location (e.g. in the map above with five clusters of the nebuloides group). If adopted, this classification means that any future investigation of the broad environmental differences between these species is compromised—it becomes circular to define species based on geography, and then examine differences between the species in environmental variables related to geography. Alternatively, you could sequence some DNA and figure out which lineage your new population belongs to. But if these three lineages are in fact one species under other non-phylogenetic criteria, then describing them as three distinct species messes up any future effort to understand correlates of speciation. You could look at hemipenis morphology, but then future studies looking at the reproductive outcome of matings between females and males from different hemipenis-species will be framed as investigations of hybridization or introgression between species, instead of a possible refutation of the relevance of hemipenis morphology to speciation (as described above for Anolis polylepis and Anolis osa).
The upshot is that species descriptions, and the basis on which they are made, are not neutral in terms of their consequences. They affect how research going forward is framed, how we design our sampling, and how we interpret our results, which is why I agree with you that we should all be worried about this explosion of new species. I don’t quite know what the answer is—personally, I’m fine with species that harbour lots of variation, and your mileage may vary. But going out on a limb here, what’s the harm with figuring out phylogenetic relationships between populations, sampling across the phylogeny of the group for subsequent research, and holding off on species descriptions until we have multiple, very well-developed lines of evidence that support the classification?
Martha Muñoz
I’d like to propose splitting this post into multiple posts.
Martha Muñoz
Sorry, couldn’t help myself! Great post, Levi.
Ambika Kamath
HA!
Levi Gray
Ha! It was considered, and perhaps I should have split it up. As long as this post is, there are a few related topics I was tempted to go into further. In particular, I think it would be worth discussing speciation in anoles in general–what traits (if any) are more commonly associated with divergence, what constitutes strong evidence for species-level divergence, etc.
I’ll try to make the next one a more reasonable length.
Levi Gray
One thing worth adding to my comments about dewlap color–a recent paper in BMC Evolutionary Biology by Julienne Ng, Alison Ossip-Klein, and Rich Glor has challenged the assumption that divergence in dewlap color corresponds with reproductive isolation. The study is really worth checking out:
DOI: 10.1186/s12862-016-0763-4
Anthony Geneva
Levi, nice job kicking off an important and often avoided discussion.
My perspective comes from someone who focuses on studying the process of speciation rather than diagnosing species. I’m interested in the underlying causes of speciation, focusing on the evolution of reproductive isolation. Although readers of Anoles Annals may disagree on their favored species concept, I don’t know of any species concept that would reject reproductive isolation as evidence of species status. Despite this, we rarely test for reproductive isolation. I would love to advocate for everyone to cross their putative species to directly measure intrinsic reproductive isolation but carrying out such experiments in anoles is not easy (to be honest, it is really hard).
It is far more common for researchers to look for patterns which may suggest morphological, genetic, or reproductive discontinuities. The absence of intermediate states between discontinuities then serves as evidence that populations are not interbreeding due to some (usually unknown) reproductive barrier. For instance, reciprocal monophyly between samples drawn from different populations is often used as evidence of species status but speciation is not the only path to reciprocal monophyly. Rather, cessation of gene flow between populations leads to coalescent processes eliminating shared ancestral variation and, given sufficient time, results in reciprocally monophyletic populations. This pattern does not tell us that these populations are species, only that populations have been separated long enough for coalescence to occur in the markers sampled. For folks not familiar with coalescent theory, this can happen rather quickly, particularly when studying small populations or when using uniparentally inherited loci (like mitochondria used in the studies that Levi mentioned). Reciprocal monophyly often occurs between groups that remain fully interfertile (e.g. are still a single species). Similarly, differences in the number of subdigital lamellae or the size, shape, and number of scales don’t cause speciation, we study such traits because they can arise as a consequence of speciation.
Since the traits we often study are outcomes of the speciation process, it can be appealing to try to find traits that simultaneously allow us to distinguish species AND actually serve as a reproductive barrier. As Levi mentioned, recent work has focused on hemipenis morphology in anoles for just this reason. The idea here is that this trait can not only diagnose species, it may actually represent a reproductive barrier between populations by way of a “lock-and-key” mechanism. The lock-and-key hypothesis states that divergence in genital morphology between populations of internally fertilizing animals can result in males being physically unable to transfer sperm to females of other populations, resulting in reproductive isolation. This theory enjoyed particularly strong appeal among groups studying arthropods where chitinous reproductive anatomy suggests that divergent structures may be physically incapable of intromission. More recently, the structural hypothesis has lost favor even among arthropods as the evidence that divergent reproductive structures do not prevent sperm transfer (reviewed in http://dx.doi.org/10.1155/2012/247352). If lock-and-key fails in the rigid genitalia of arthropods it should come as no surprise that hemipenis morphology is not a strong barrier to introgressive hybridization in anoles.
In the absence of such dream traits (those that tell us what species are and how they are maintained), we are left only with distributions of traits that might suggest reproductive isolation. If this is the case, I argue that the bar must be set high for this type of correlative evidence. Taxonomic decisions are hypotheses that generate testable predictions. In my view, well-constructed taxonomic hypotheses should be built upon strong evidence, where discontinuities are explicitly described and good faith efforts have been made to detect intermediate or shared of diagnostic traits. Selecting to study scattered sites across a continuous range without sampling in-between can directly lead to positively misleading species diagnosis. I think Levi and Ambika made it clear that not only do such incorrect diagnoses waste the community’s time, it also has the potential to mislead analyses that rely on accurate assessment of species boundaries.
Gunther Koehler
>>> Gunther Koehler 15.10.16 12.33 Uhr >>>
>>> Gunther Koehler 15.10.16 12.11 Uhr >>>
I am not a big blogger but since Levi’s blog relates directly to some of my work and because his blog contains several misleading statements and false assumptions, I feel compelled to drop a few lines. Levi states that he has observed “hemipenial polymorphism within species”. I have everted several thousand anole hempenes over the past 25 years and have not yet observed hemipenial polymorphism in any species at a single locality. Despite its soft tissue nature, variation in hemipenial morphology is negligible in specimens from the same locality and within species limits over some considerable geographic distances (up to a few thousand kilometers in species such as capito and biporcatus). In case two hemipenial morphs are present at a given site, then this indicates the presence of two species. And then again, you will find two discrete morphs and not intermidiate forms.
However, although the study of hemipenial morphology is pretty straight forward, there are some caveats. In particular, it is essential to consider only hemipenes of ADULT males that are COMPLETELY EVERTED. There is ontogenetic variation in some species (most evident in the relative length of the lobes), and it is useless to study incompletely everted organs. That said, I assume that the “hemipenial polymorphism within species” observed by Levi is simply the result of including incompletely everted organs, and those of subadult males and/or the mixup of more than one species. In a recently submitted manuscript by Levi and coauthors that I reviewed, he had clearly lumped together several species under a single name, which led him to believe he had more than one hemipenial morph in “species” he studied. Of course, he had found discrete hemipenial morphs and no intermediate states. Hemipenial morphology obviously is not selectively neutral and in our lab we found that the female cloaca coevolves in species pairs with divergent hemipoenial morphology, having muscular vaginal tubes in species where the males have bilobed hemipenes, and very reduced or no such tubes in the unilobed species.
I never stated that there is a “lock and key” mechanism at work in anole copulatory organs. The divergent hemipenes actually are no good species barriers. In the lab, we unintendedly produced hybrids between distantly related species such as polylepis and humilis! However, hemipenis morphology is a strong and very useful tool in anole taxonomy. It might not solve every problem – but which method is universially useful? At this point, to my knowlegde, no compelling evidence for “hemipenial polymorphism within species” has been published. Thus, dear Levi, please provide evidence and – until then, refrain from distributing false information on an important topic. Way too few anole researches study hemipenis morphology in lizards, and what we least need, is to discredit a powerful taxonomic tool.
Levi Gray
First of all, thanks for taking the time to comment.
One of my concerns is that if you diagnose species by hemipenes, you can never find polymorphism within species using hemipenes. Furthermore, I think the best published evidence for hemipenial polymorphism within species is your research.
Hemipenes were the only evidence cited for recognizing Anolis osa as a species (Kohler et al. 2010). I’m attaching two figures from the followup paper that looked into the polylepis/osa contact zone (Kohler et al. 2012). The first is the mtDNA tree, with individuals labeled “os” for osa, “p” for polylepis, and “h” for hybrid.
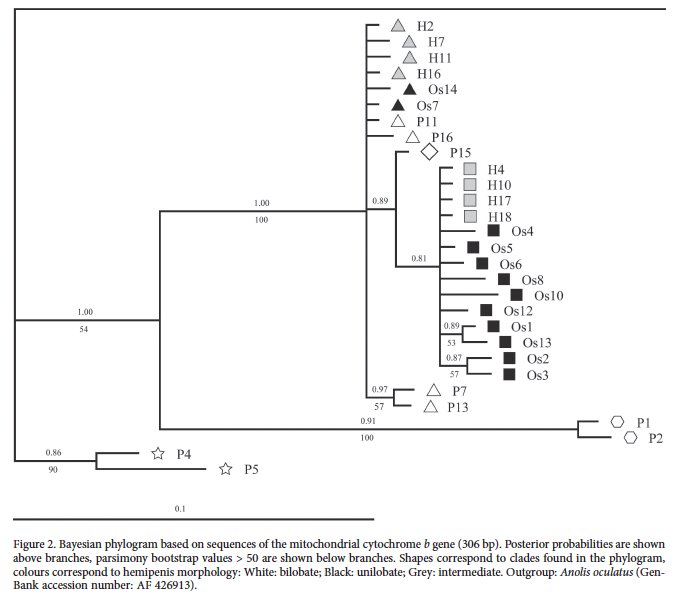
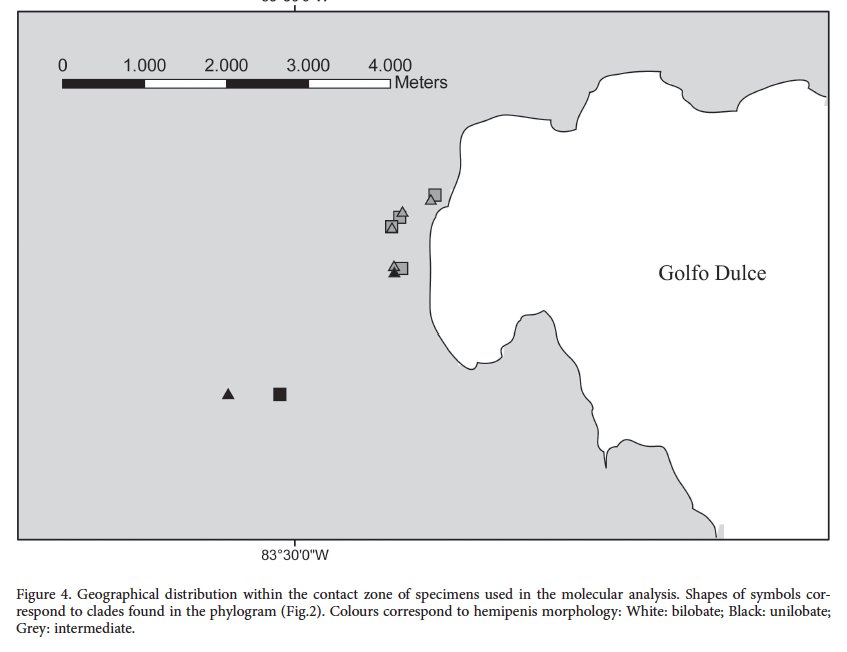
The second is the map of sampling localities.
It is not a stretch to consider this paper a good example of hemipenial polymorphism in anoles. We have no evidence of reduced gene flow and it appears the populations, diagnosed by hemipenes, are continuous and freely interbreeding. I am not aware of a currently accepted species concept utilized in vertebrates that would support the validity of Anolis osa as a species. The morphological and molecular data presented supports the presence of a single species that consists of at least two geographically distributed subpopulations that are often diagnosable by hemipenial morphology.
You also say that the lack of intermediate hemipenial types is evidence that, in other systems, populations with different hemipenes are valid species. I don’t agree with that sentiment in general, as there are many examples of traits (with dominant alleles, for example) where intermediate forms of traits do not exist. But in this particular case (osa/polylepis), there are intermediate forms. You and your co-authors still came to the conclusion that they should be recognized as separate species. I do not understand this interpretation of the data.
I would also like to point out that in no way am I suggesting that hemipenial morphology is useless or neutrally evolving in anole evolutionary studies. There are some very promising avenues of research involving anole hemipenes. As I have expressed in the above blog, I am worried that the way these traits have been utilized in recent taxonomic research is not justified. And the cost of using these traits inappropriately to split up populations that are not valid species is high. Due to the high cost of taxonomic mistakes, changes should not be taken lightly and should require significant evidence.
Levi Gray
The map above is zoomed in on the contact zone region, and does not show where the rest of the samples were from. But it does make clear that there are multiple hemipenial morphs present in the same area and no molecular differences detected between the two putative populations in the tree.
Ambika Kamath
What I struggle to understand is why hemipenis morphology is considered a valuable taxonomic tool if it is entirely divorced from reproductive isolation. If the advantage of quantifying hemipenal morphology is that it correlates with the degree of genetic divergence, then is it worth considering it a separate line of evidence when delimiting species? Does this in turn mean that we are adhering to a purely phylogenetic species concept, and if so, how much divergence is enough to describe a new species? I apologize if these questions are ignorant from a taxonomic standpoint, but given the extent to which species descriptions matter to non-taxonomists, I think it is important that we all understand them.
Gunther Koehler
Levy, it is amazing what you read out of our 2010 paper on polylepis and osa! In that article, we actually provided evidence that ALL specimens from the mainland (n = 161 males) throughout the geographic distribution area of polylepis have a bilobed hemipenis – stretching a vast area of some 300 km in NW-SE direction and up to 100 km distance towards the Talamancan mountains. Not a single male from this area with a unilobed hemipenis among the numerous specimens. In contrast, ALL males (n = 58 males) from the Osa Peninsula south of the village of Rincon de Osa had a small, unilobed hemipenis – no variation whatsoever! Just look at Figure 6 of our 2010 paper. But we detected a hybridization zone of about one kilometer width just near the village of Rincon de Osa; and in this narrow strip we found males with an intermediate hemipenis morphology – just as expected for hybrids! Thus, again we look at a strong geographic correlation of hemipenis morphology and no polymorphism! Show me a unilobed polylepis from the mainland and we can talk about evidence for polymorphism …
Levi Gray
Without a significant reduction in gene flow between the two subpopulations, you are describing geographic variation within species. It does not matter if you can diagnose the subpopulations consistently by hemipenial morphology if you demonstrate no reproductive isolation between those populations.
What we know from you and your colleagues’ research is that there are subpopulations that run into one another near the village of Rincon de Osa. These subpopulations can apparently be diagnosed by hemipenial morphology and nothing else. You then demonstrated that there appears to be uninterrupted gene flow between the two subpopulations.
The only justifiable reason to elevate those hemipene-diagnosed subpopulations to species status is if the lineages are experiencing reproductive isolation of some sort. Otherwise they are, by any widely-accepted species concept, conspecific.